Human genetic mapping can provide insight about COVID-19 pathogenesis and drug development
Published: 2021-08-10
Over the last two years, the COVID-19 pandemic (caused by the SARS-CoV-2 virus) has burdened healthcare systems and economies worldwide. Treatments and vaccines against COVID-19 have been developed in record time over this period. The effects of SARS-CoV-2 infection on patients have been found to vary from asymptomatic infections to life-threatening conditions, including viral pneumonia and acute respiratory distress syndrome. Multiple studies have identified risk factors (e.g. age, sex, and body mass index) that can influence disease severity, but these risk factors alone do not explain disease variability between individuals. Investigations into how genetic factors may contribute to COVID-19 susceptibility and disease severity may therefore be crucial in providing important insights into COVID-19 pathogenesis and identifying potential mechanistic targets for drug development or drug repurposing.
Previous genome-wide association studies (GWAS) have provided evidence for the involvement of several genomic loci (e.g. 3p2) in COVID-19 severity and susceptibility. Additional understanding regarding the genetic basis of individual susceptibility to SARS-CoV-2 and severity of COVID-19 disease is needed.
A recently published study in Nature (accepted June 23rd 2021, published July 8th 2021) by the COVID-19 Host Genetics Initiative (HGI) (a global research network) reports the results of meta-analyses of 46 COVID-19 host genetic effects studies from 19 countries. The study includes results from the Swedish SweCovid study, which focused on genetic susceptibility to the development of severe COVID-19 symptoms. Researchers from both Uppsala University (Michael Hultström, Miklós Lipcsey, and Robert Frithiof) and Karolinska Institute (Hugo Zeberg, Jonathan Grip, and Nicolas Tardif) have contributed to the SweCovid study.
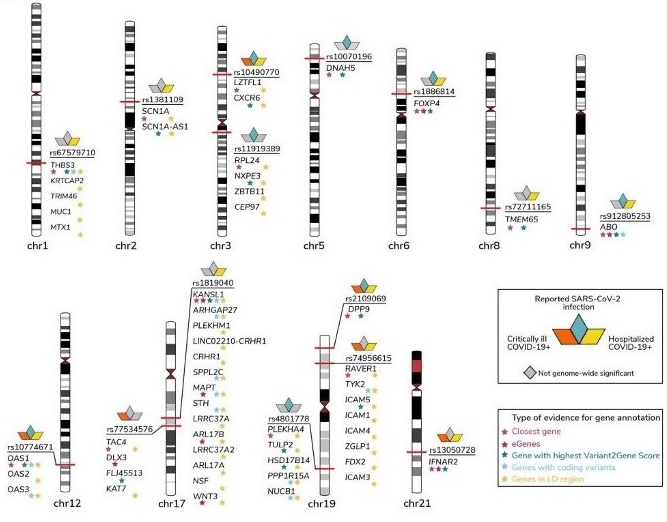
In their study, researchers in the COVID-19 Host Genetics Initiative describe the results of three genome-wide association meta-analyses comprising 49,562 COVID-19 patients and almost two million unaffected individuals. To be included in the meta-analyses, studies had to involve a minimum of 50 cases and 50 controls in the analysis. The meta-analyses identified 13 genome-wide loci that are associated with SARS-CoV-2 infection or severe manifestations of COVID-19. Of the 13 loci, six were associated with the development of critical illness due to COVID-19 (based on 16 studies), and nine loci were associated with moderate to severe COVID-19 (based on 29 studies). In addition, seven of the 13 loci were associated with SARS-CoV-2 infection. Notably, no sex-specific effects were detected, despite males being at greater risk of severe disease. Interestingly, two of the loci are close to the previously reported region of interest in the 3p21.31 region of chromosome 3. Further, a number of the loci identified in the study have previously been found to be associated with lung or autoimmune, and inflammatory diseases; these also represent potentially actionable mechanisms in response to infection.
The study highlights the importance of including data from diverse populations for genetic discovery. In particular, two of the 13 loci were shown to have higher allele frequencies in individuals with South East Asian (rs1886814, 15%) and East Asian genetic ancestry (rs72711165, 8%), compared to those with European ancestry (< 3%).
The researchers completed Mendelian Randomisation analyses to examine the effect of risk factors. The results support a causal role for smoking and BMI in the development of severe COVID-19 symptoms, but the association with type II diabetes was not confirmed.
“I think it is very exciting that we have been able to contribute to this massive international effort during the ongoing pandemic. It opens up new venues of research that can allow us to reduce the number of patients that get critically ill.” says Miklós Lipcsey, professor of Anaesthesiology and Intensive Care Medicine at Uppsala University.
In conclusion, the study represents one of the largest genome-wide association studies to date. This study underlines the importance of international collaboration; by combining a large number of studies from 19 counties in the meta-analyses of GWAS data, the research initiative was able to make important discoveries about the role of human genetics in SARS-CoV-2 infection and COVID-19 disease severity. These results may contribute to future studies about the complex new SARS-CoV-2 virus and COVID-19 disease severity, and could also be used as a model for pandemic preparedness.
In order to increase FAIRness, the research initiative shared a preprint of the study via the MedRxiv preprint server in the spring of 2021. Harmonised individual level data is deposited at the European Genome-Phenome Archive and can be accessed with appropriate permissions. The code used in the study for producing summary statistics, the projection PCA pipeline (including precomputed loadings), and meta-analysis is available on GitHub. The code for Mendelian randomisation and the genetic correlation pipeline is available on a separate GitHub Repository.
The SweCovid study was supported by SciLifeLab National COVID-19 Research Program and Knut and Alice Wallenbergs stiftelse, the Swedish Research Council the European Union’s Horizon 2020 research and innovation programme, the Hjärt och Lungfonden, Njurfonden, and Nordforsk.
For more information about the SweCovid project, see the SweCovid website. Previous Data Highlights based on this study can be found here: Data-driven research shows that Neanderthal gene connected to severity of COVID-19, and Neanderthal gene protecting against severe COVID-19 could be potential new drug target.
Data
- Harmonised individual level data: European Genome-Phenome Archive
- Code for producing summary statistics, the projection PCA pipeline (including precomputed loadings), and meta-analysis: GitHub
- Code for Mendelian randomisation and the genetic correlation pipeline: GitHub
Article
DOI: 10.1038/s41586-021-03767-x
COVID-19 Host Genetics Initiative. Mapping the human genetic architecture of COVID-19. Nature (2021)